|
Current Level |
|||||||
|
|
|||||||
|
Previous Level |
|||||||
|
|
|||||||
|
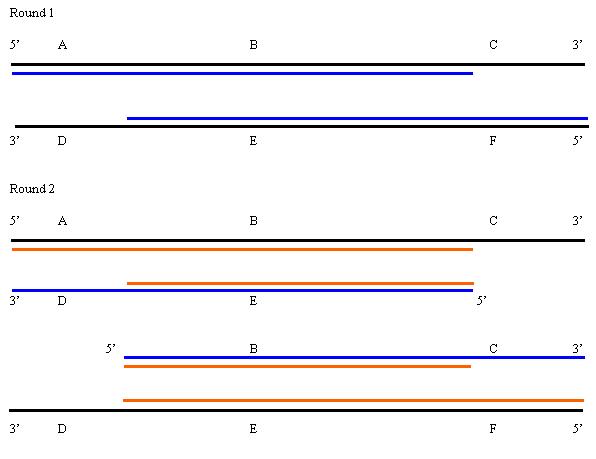
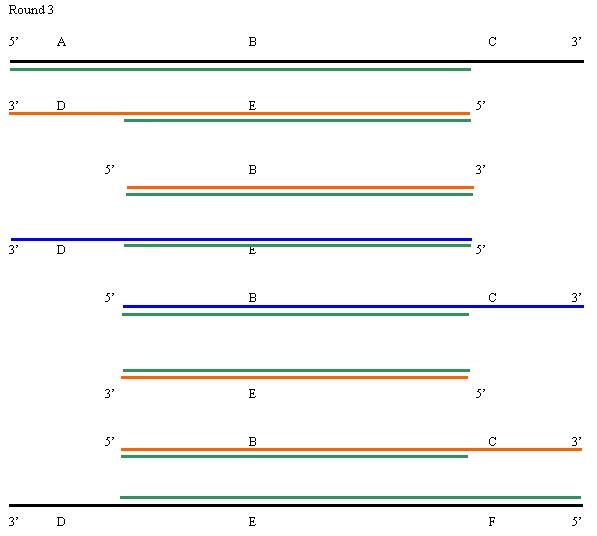
1. Each round of replication leads to a doubling of the amount of DNA in the region between the primers. However, the final PCR product does not appear until the third round of amplification. |
 |
||
 |
||
|
2. First determine the reverse complent sequence to this strand of DNA. Then design primers (shown in red) that will bind to the 3´ end of each strand of DNA. 3´ GGCATCGACCTCCCT 5´ 5' GGATCGATCAAGAACAATGACAGGATCGAGGAATTCAGCCTACGCAGCCCGTAGCTGGAGGGA 3'3´ CCTAGCTAGTTCTTGTTACTGTCCTAGCTCCTTAAGTCGGATGCGTCGGGCATCGACCTCCCT 5´ 5´ GGATCGATCAAGAAC 3´ What other reagents are necessary to perform a PCR reaction?Taq polymerase, nucleotides (dNTPs) and a buffer PCR machines cycle between three temperatures. What is the purpose of each stage in the PCR cycle, and roughly what temperatures are used?
Product is formed in the third round, 2 molecules of product (=21). After that it should double every round for the next seven rounds. Thus you would expect a total of 28=256 molecules. Note that you will continue to make more of the heterogeneous products each round, so this is really an underestimate.
|
|
|
|