![]()
In April of 2001 at the University of Heidelberg, Germany, a team of scientists led by Dr. Richard Hermann finished work sequencing the genome of M. pneumoniae. This work was important because M. pneumoniae is thought of as a model “minimalist cell”, meaning the DNA in the bacteria only codes for proteins that are absolutely obligatory for survival. All the information for M. pneumoniae is encoded on a lone circular double strand of DNA.
The entire genome for M.
pneumoniae is only 816,394 base pairs long, coding for 689 proteins
total. Compare this to the genome of another bacterium, Escherichia coli,
which is 5,082,025 base pairs long and codes for 4,573 proteins. The genome
of M. pneumoniae is so small, it cannot synthesize it own purines (Adenine
and Guanine, two of the nucleotides that make up DNA and RNA molecules.)
The Mycoplasms are thought to have evolved from more complex gram positive
bacteria. Somewhere through evolution, M. pneumoniae lost the ability to encode
for a cell wall, purines, and many proteins. Purines include
Adenine and Guanine, which make up about 60% of the M. pneumoniae genome. This
and the lack of a cell wall, is why M.
pneumoniae are obligate parasites of human cells, where they can attain
necessary amino acids and phosphates needed for survival. 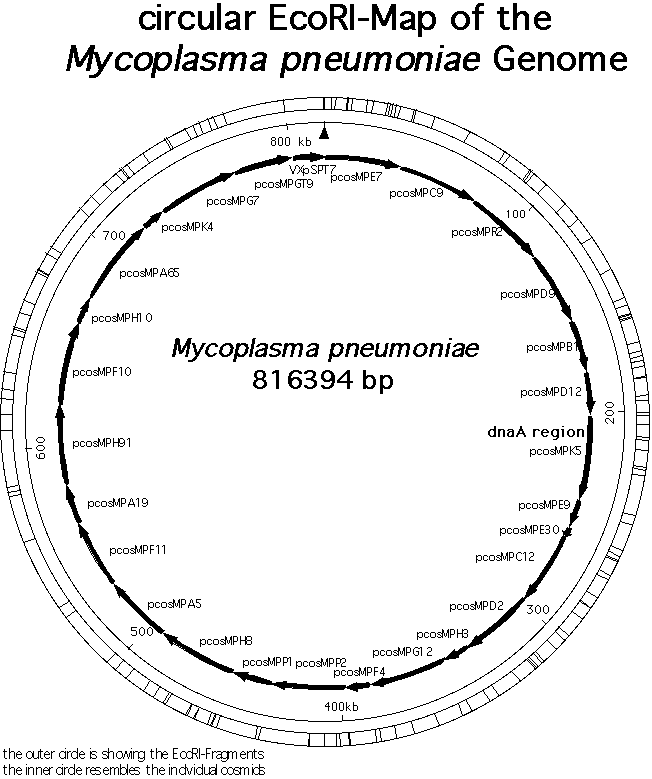
Genome of Mycoplasma pneumoniae