|
Current Level |
||||||||||||
|
|
||||||||||||
|
Previous Level |
||||||||||||
|
|
||||||||||||
Answers to Short Answer QuestionsMOLECULAR SWITCHES IN THE LIVER We have seen how RNA polymerase and general transcription factors assemble on a promoter to transcribe a mRNA. In this section we will explore how this process is regulated by the DNA itself and other transcription factors and signalling molecules.
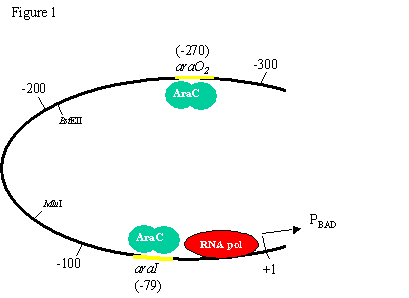
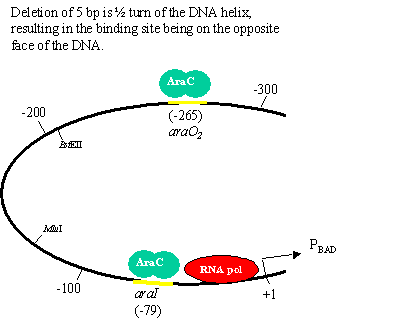
Role of DNA and chromatin structure One question that puzzled many scientists in the 1980s was how proteins binding to one region of DNA could regulate transcription of genes 100s-1000s of bases away. This paper elegantly addresses this question. (Note: only portions of the paper are shown) 1. What questions are the authors trying to answer about size limits and helical repeats? What is the general design of their experiment? The authors are trying to determine whether the distance between a DNA binding site an the promoter is important in regulating transcription. They also want to use this to predict the actual periodicity of DNA in vivo. 2. How do the authors increase the distance between the two AraC binding sites (araI and araO2)? What is the purpose of the enzyme BstEII? The E. coli DNA fragments? The short oligonucleotides? The authors cut the region between the two AraC binding sites with BstII and inserted randomly sized DNA fragments to increase the distance between the sites. Short oligonucleotides were used to make more specific increases in distance to determine periodicity. 3. How did the authors decrease the distance between the two AraC binding sites (araI and araO2)? What was the reason for using BAL-31 exonuclease? The authors cut the region between the two AraC binding sites with BstII and then added an exonuclease to chew away bases. The plasmid was then religated, resulting in a series of deletion mutations. 4. When AraC binds to both the araI and araO2 sites what affect does this have on the araCBAD promoter (PBAD)? AraC is a repressor, so expression is repressed. 4. What is the average number of bp per turn in normal DNA in solution? What periodicity do the authors find in vivo? In vitro = 10.5 bp/turn In vivo = 11.1 bp/turn 5. What interesting observation do the authors make in figure 3? Using this data, suggest a mechanism by which proteins binding to one region of DNA could regulate transcription of genes 100s-1000s of bases away. (Hint: look at figure 1). In addition to the difference in periodicity, the authors could explain how proteins binding to sites at distances of 100s or 1000s of bp from a promoter could regulate transcription.
Proteins regulating transcription One way to study the role of a gene in an organism is to delete the entire gene in a mouse and examine the phenotype of the animals. Such animals are call transgenic animals, or gene knock-outs. We will go into how this is done later, for now assume that when they say genotype +/+ they mean a mouse with both copies of the indicated gene, +/- is a heterozygote and -/- is a homozygous mouse which has lost both copies of the indicated gene. Null ES cells refer to cells from these mice. Article: Regulation of a Transcription Factor Network Required for Differentiation and Metabolism Stephen A. Duncan, * M. Angeles Navas, Daniel Dufort, Janet Rossant, Markus Stoffel. Science, 281:5377, 692-5, (1998) 1. RT-PCR is reverse transcriptase PCR. What will the authors be detecting using this technique?
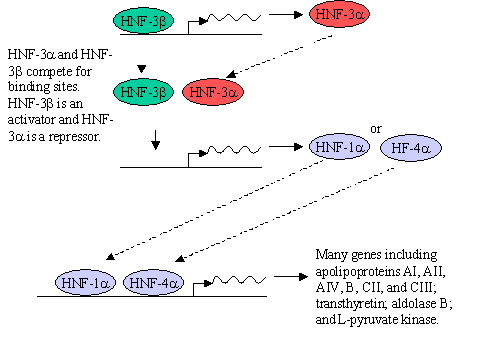
What type of genes are being knocked out? Hepatocyte Nuclear Factors (HNF) are transcription factors What is a hepatocyte? A liver cell. 2. In figure 2 which gene has been knocked-out? Which genes appear to be regulated by this protein? Does it appear to be a positive or negative regulator? HNF-3 3. In figure 3 which gene has been knocked-out? Which genes appear to be regulated by this protein? Does it appear to be a positive or negative regulator? HNF-3 4. Comparing the HNF-3 a and HNF-3b bands in both figures 2 and 3, explain the presence or absence of each band in the (-/-) mice.HNF-3a is missing in Fig. 2 because it has been knocked out. HNF-3b is not affected because its expression is not regulated by HNF-3a. HNF-3b is missing in Fig. 3 because it has been knocked out. HNF-3a is also missing because its expression is regulated by HNF-3b. 5. Draw a diagram of how HNF-3a and HNF-3b may regulate the expression of genes in a cell. HNF-3b regulates the production of HNF-3a. Both can bind to the promoters of HNF-1a and HNF-4a. HNF-3a is a repressor and HNF-3b is an activator of HNF-1a and HNF-4a transcription. In turn, HNF-1a and HNF-4a are transcription factors that regulate the production of many liver proteins.
|
|
|
|