|
Current Level |
||||||||||||
|
|
||||||||||||
|
Previous Level |
||||||||||||
|
|
||||||||||||
Regulation of Transcription (lac operon)Much of the original work deciphering how transcription is regulated was done on the lac operon. The 1965 Nobel Prize in Medicine was awarded for this research. Bacteria often organize genes in groups with similar functions called operons. RNA polymerase binds to the promoter at the 5' end of the operon and transcribes the genes into RNA. The 2006 Nobel Prize in Chemistry was awarded for research on the structure and regulation of RNA polymerase. Bacteria need to be very efficient, and only express enzymes when they are needed. An example is in metabolism of lactose, a disaccharide found in milk. Bacteria will express the enzyme beta-galactosidase the highest when they only have lactose as an energy source. If other energy sources such as glucose are present, expression of beta-galactosidase will be decreased. Finally, if no lactose is present at all, beta-galactosidase will not be expressed.
Transcription and TranslationFor a bacteria to express the genes on the lac operon they are first transcribed by RNA polymerase. The regulation of the expression of most genes is done at the level of transcription. The transcribed mRNA is then translated into proteins by ribosomes.
Response to LactoseE. coli express a protein called the lac repressor. In the absence of lactose, this protein can bind to DNA near the promoter for the lacZ gene. This prevents RNA polymerase from binding, and no transcription is possible.
If lactose is present, the lactose binds to the lac repressor, inducing a conformational change that causes the repressor to fall off of the DNA and transcription can proceed at a fairly low level..
Response to GlucoseLow levels of glucose levels in a cell lead to the formation of cyclic AMP (cAMP). cAMP binds to a protein called CAP or CRP. Once CAP is bound to cAMP, it can bind to a region upstream of the promoter, where it accelerates RNA polymerase binding. This leads to increased transcription of the lacZ gene.
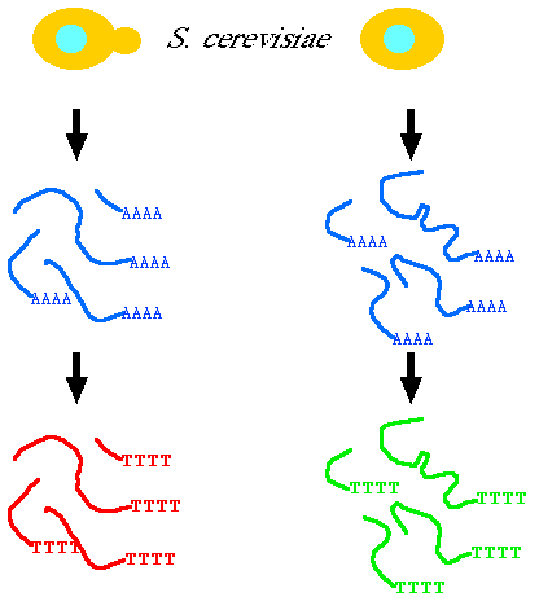
RNA ProcessingEukaryotic genes contain Exons (which encode information that will end up being translated into protein) separated by Introns (sequences that will not encode proteins). At the 5' end of a gene we find a promoter and often a CpG island. Both of these elements regulate the transcription of a gene. At the 3' end of a gene we find a stop sequence, and a signal for polyadenylation (AAUAAA). Transcription is the process of copying the genetic information in DNA into RNA. This is done by the enzyme RNA polymerase which binds to a region called a promoter found at the 5´ end of a gene. The binding of RNA polymerase is tightly regulated by many proteins called transcription factors. RNA polymerase transcribes the genomic DNA sequence into the corresponding RNA sequence resulting in the formation of a primary transcript. In Eukaryotes this primary RNA transcript is then processed into a messenger RNA (mRNA) if the gene is to be translated into protein. This involves removing introns, which do not encode for protein, and splicing the remaining exons together. At the 5´ end of the mRNA a 7-Methyguanosine residue is added to provide a protective cap. At the 3´ end of the mRNA, 50-100 adenosine residues are added, generating a poly A tail. |
|
Genomic DNA
Primary Transcript (RNA
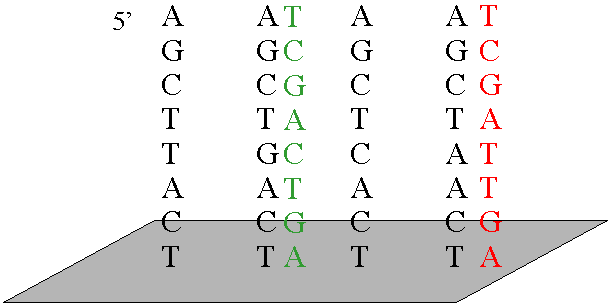
Messenger RNA (mRNA) Measuring TranscriptionTo measure transcription we can follow mRNA production. This has traditionally been done with Northern blots or reporter genes. Now techniques like Reverse Transcriptase PCR (RT-PCR) and Microarrays are commonly used. In a microarray, either mRNA is spotted onto a glass slide, or DNA is synthesized onto a chip. mRNA samples are then isolated from cells, and fluorescently labeled by RT-PCR.
The labeled cDNAs are then added to the glass slide or DNA chip, and hybridization is detected by the relative amount of fluorescence.
In this way, the transcription of thousands of genes can be measured simultaneously. Practice Problems |
|
|
|