|
Current Level |
||||||||||||
|
|
||||||||||||
|
Previous Level |
||||||||||||
|
|
||||||||||||
|
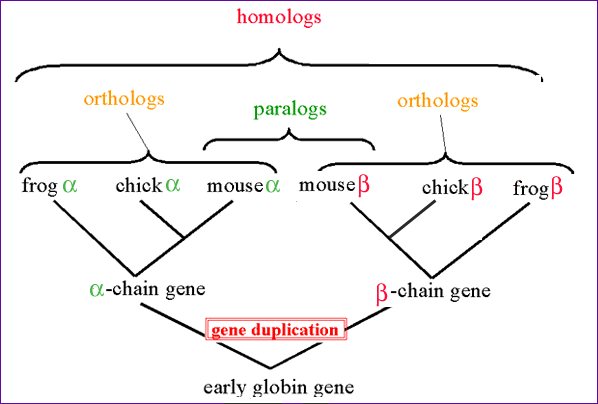
Protein Families We have seen one way to make predictions about a protein based upon its primary sequence by looking for specific motifs or patterns that would predict things like activity, secondary structure or hydrophobicity. Next we will compare a protein´s sequence to that of other proteins to see if it belongs to a family of proteins. Proteins are grouped into families based upon similarities in structure and function, and are thought to have evolved from a common ancestoral protein through gene duplication and subsequent mutation. The SCOP database (Structural Classification of Proteins http://scop.mrc-lmb.cam.ac.uk/scop/) groups proteins by family and superfamily. You can search this database by keyword, or browse by family. When we talk about related proteins we use specific terminology. Homologs: are sequences that have common origins but may or may not have common activity.Orthologs: are homologs produced by speciation. They represent genes derived from a common ancestor that diverged due to divergence of the organisms they are associated with. They tend to have similar function. Paralogs: are homologs produced by gene duplication. They represent genes derived from a common ancestral gene that duplicated within an organism and then subseqeuntly diverged by accumulated mutation. They tend to have slightly different functions. From Bioinformatics, Baxevanis and Ouellette, 2nd Edition, 2001, p. 327, Wiley Pub.
From the Stanford Folding@home glossary
www.stanford.edu/.../
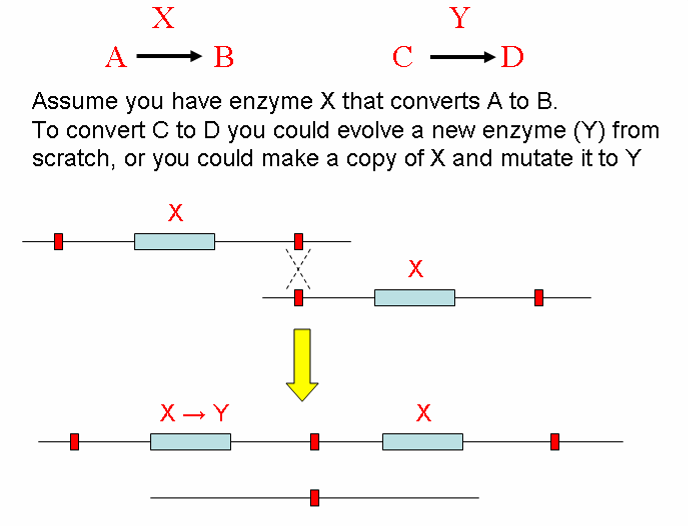
How do new proteins arise?
Gene Duplication Provides Template for New Proteins to Evolve
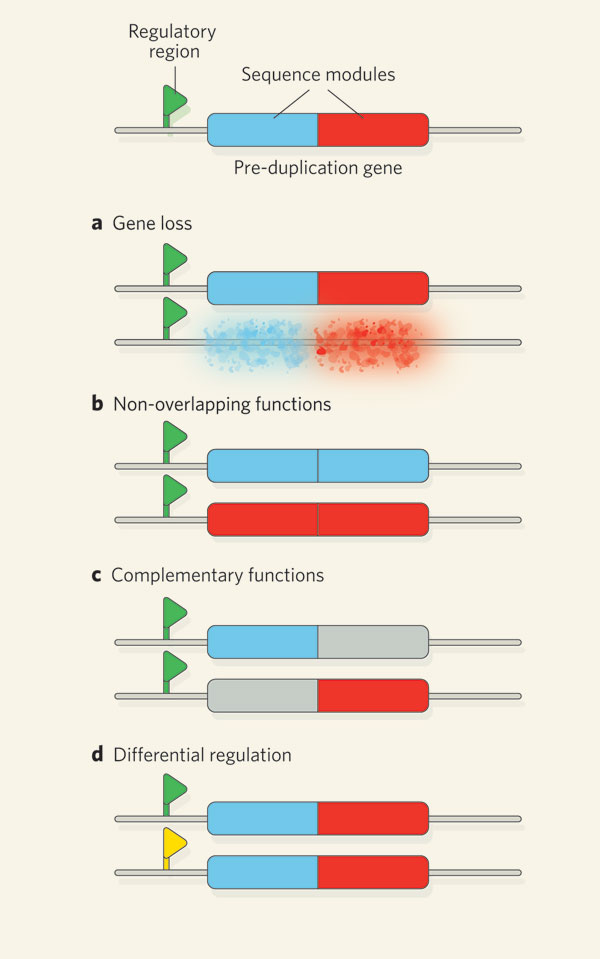
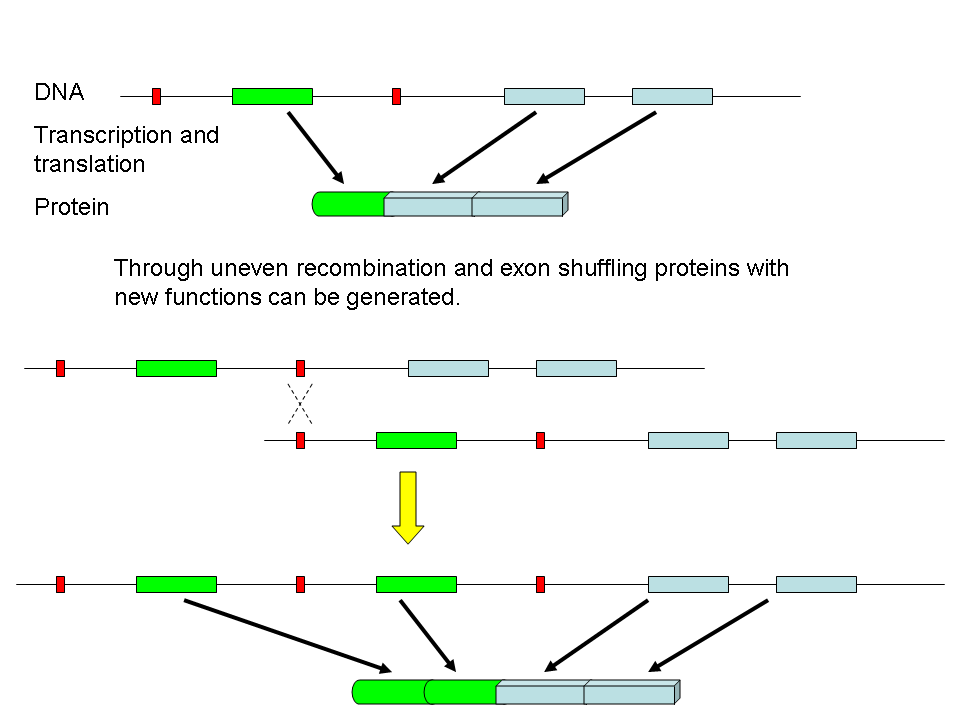
Domains are often carried on exons.
In addition, we can change how we splice mRNA. This gives different combinations of proteins from one mRNA.
By swapping domains a proteins activity can be changed New proteins are made by exchange of domains and by mutations within domains Add additional domains or make mutations to change members of the same family. Paralogs or Protein Families - Serine Proteases Next we will examine a specific family of proteins. Enter BIOLOGY WORKBENCH and find the sequence we used for trypsin last week under Protein Tools. To find related proteins we will use a BLASTP search. Select the database H. sapiens proteins for the search. Select 6-7 sequences from the results of this search. If you hold down the Ctrl key you can select multiple individual sequences. At this point, only select human protein sequences. Try to pick a variety of sequences, i.e. some that are closely related and some distant family members. Don't go below a score less than 100, or the alignment starts to fall apart. Import these sequences into Biology Workbench Align these sequences using CLUSTALW. Be sure to also align them with both the SWISSPROT and PDBFINDER sequences that your assigned sequence matched with. Examine the alignment and phylogenetic tree. Import your alignment and view it with BOXSHADE (you can also save this as a figure for your report).
Go back to the original Biology Workbench window and select Protein Tools. Select the other serine protease sequences and analyze them with PROSEARCH.
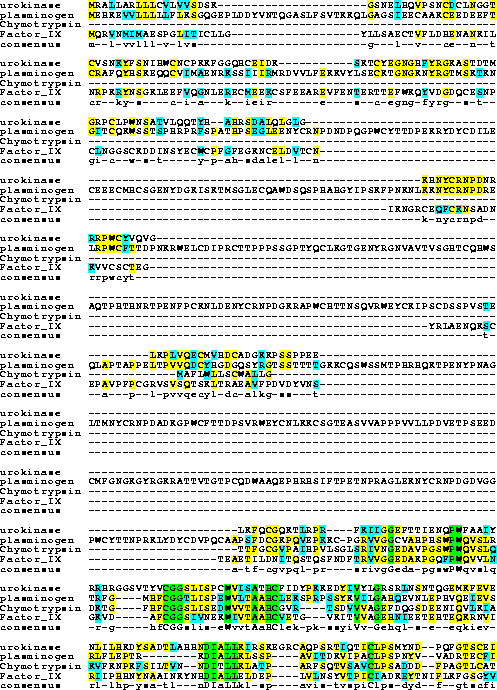
Motif Search (Link to Biology Workbench) Compare Urokinase, Factor IX and Plasminogen
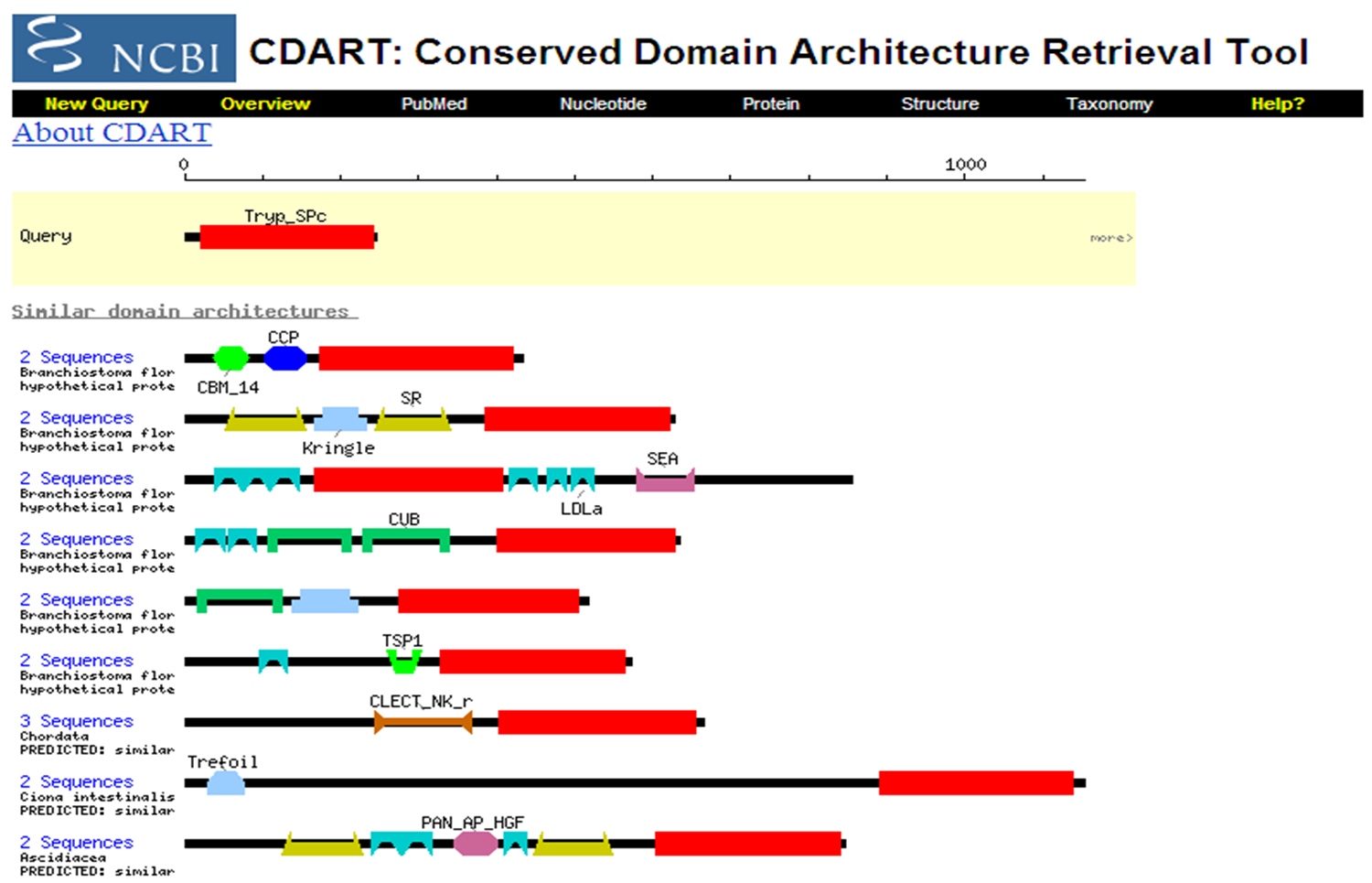
DART Domain Architecture Retrieval Tool Examine Plasminogen (Acession # P00747) Click on each domain to learn its function Click on ``28 similar domain architectures" This will display orthologs and paralogs. What is the major difference between some of these proteins? At the bottom of the page click on ``Next". There are 10 pages of proteins that contain at least one of the domains in Plasminogen. These domains have been combined with other domains to create unique proteins.
Orthologs - Serine Proteases Next we will examine trypsin orthologs. Enter BIOLOGY WORKBENCH and find the sequence we used for trypsin last week under Protein Tools. To find related proteins we will use a BLASTP search. Select the databases GenBank Mammals, GenBank Invertebrates, GenBank Fungi, GenBank Bacteria for each search. Do each search separately, or it will be difficult to find some of the more distant matches as they will be hundreds of lines down in the results. Select 6-7 sequences from the results of this search. If you hold down the Ctrl key you can select multiple individual sequences. Be sure you are picking the same protein in the different species, and try to get a variety of species. You may not be able to find your protein in all species, for example, trypsin is in invertebrates like drosophila, but not plants, fungi or bacteria.. Import these sequences into Biology Workbench Align these sequences using CLUSTALW. Be sure to also align them with both the SWISSPROT and PDBFINDER sequences that your assigned sequence matched with. Examine the alignment and phylogenetic tree. Import your alignment and view it with BOXSHADE (you can also save this as a figure for your report).
|
|
|
|