Classification
Domain: Bacteria
The domain bacteria is characterized by prokaryotic, single celled
organisms. Because these cells are prokaryotic, they lack a true nucleus
and instead have a nucleoid region which contains free floating DNA.
These cells undergo rapid growth and cell division which can cause
them to quickly adapt to their environment. Bacteria include many
human pathogens including
anthrax (Bacillus
 anthracis), syphilis (Treponema pallidum),
Streptococcus, and
botulism (Clostridium botulinum).
anthracis), syphilis (Treponema pallidum),
Streptococcus, and
botulism (Clostridium botulinum).
Kingdom: Eubacteria
The kingdom Eubacteria are known as the “true bacteria”. Many
antibiotics such as streptomycin and tetracycline are derived from eubacteria.
Salmonella (Salmonella enteritidis) is also classified
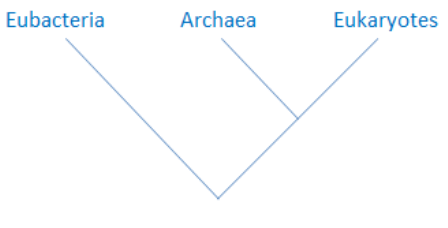
with the Eubacteria. As the above image on the left shows, all other
organisms share a common ancestor with the Eubacteria.
Phylum: Proteobacteria
These bacteria are gram-negative. This means that the outer membrane
is composed primarily of lipopolysaccharides and underneath is a
thin layer of peptidoglycan. Because of this, the gram-negative
bacteria are resistant to many antibiotics including penicillin
because they cannot get through the outer membrane. On a gram-stain,
these bacteria will appear pink-red because the peptidoglycan does
not absorb the crystal violet dye as it does in gram-positive
bacteria.
E. coli and
Neisseria Meningitidis are other examples of proteobacteria.
Class: Gammaproteobacteria
This class of organisms have a wide range of aerobicity, trophism (chemoautotrophism
and photoautotrophism), and temperature tolerance. These diverse
habitats allow this class of organisms to exploit a large range of
environments, including those without oxygen for cellular
respiration. It also includes different morphologies of bacteria
including rods, curved rods, cocci, spirilla, and filaments. Many
pathogens are included in this class including the bacteria that
caused the Bubonic plague,
Yersinia pestis,
Vibrio parahaemolyticus, as well as the bacteria responsible for
Legionnaire’s Disease (Legionella pneumophila).
Order: Aeromonadales
This grouping consists of anaerobic, rod-shaped bacteria.
Family: Aeromonadaceae
This family of bacteria do not form endospores or microcysts and
thrive primarily in aquatic inhabitants. Many species are pathogens
to humans, fish, and frogs. This family was recently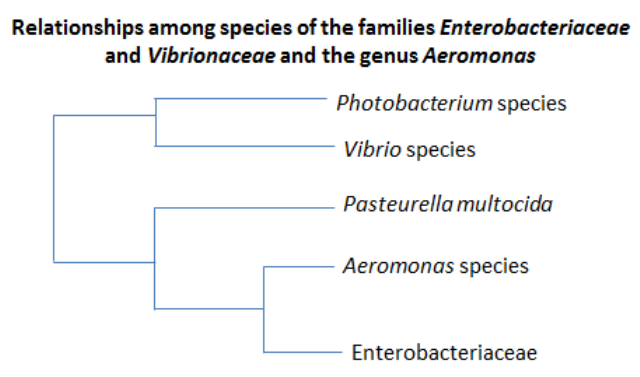 proposed in
1986 when it was found that it shared significant evolutionarily
differences from the Vibrionaceae family and that the Aeromonadaceae
are more closely related to the Enterobacteriaceae than
Vibrionaceae. The phylogenetic tree on the right represents the
evolutionary relationships based on DNA-rRNA competition data,
further showing that the genus Aeromonas is more closely related to
the Enterobacteriaceae family than the Vibrionaceae family. This
image was
redrawn from the article "Proposal to Recognize the Family Aeromonadaceae"
by Colwell, R., et al. 1986.
proposed in
1986 when it was found that it shared significant evolutionarily
differences from the Vibrionaceae family and that the Aeromonadaceae
are more closely related to the Enterobacteriaceae than
Vibrionaceae. The phylogenetic tree on the right represents the
evolutionary relationships based on DNA-rRNA competition data,
further showing that the genus Aeromonas is more closely related to
the Enterobacteriaceae family than the Vibrionaceae family. This
image was
redrawn from the article "Proposal to Recognize the Family Aeromonadaceae"
by Colwell, R., et al. 1986.
Genus: Aeromonas
This genus is characterized by their aquatic environments. A polar
flagellum allows these bacteria to be motile in these environments throughout the world including in ground and drinking water,
water distribution systems, lakes, rivers, and storage reservoirs.
Species:
Aeromonas hydrophila
 This motile species can be free-living or parasitic. It can survive
in a wide range of aquatic environments and it can also survive as a
pathogen in
an invertebrate or vertebrate host. This pathogen was found in the
floodwater following hurricane Katrina and was also the most common
cause of skin and soft tissue infections following the 2004 Thailand
tsunami.
This motile species can be free-living or parasitic. It can survive
in a wide range of aquatic environments and it can also survive as a
pathogen in
an invertebrate or vertebrate host. This pathogen was found in the
floodwater following hurricane Katrina and was also the most common
cause of skin and soft tissue infections following the 2004 Thailand
tsunami.
To learn more about its diverse habitats, click
here.
To return to the home page, click here.