|
Current Level |
||||||||||||
|
||||||||||||
|
Previous Level |
||||||||||||
|
||||||||||||
| DNA Protein Interactions |
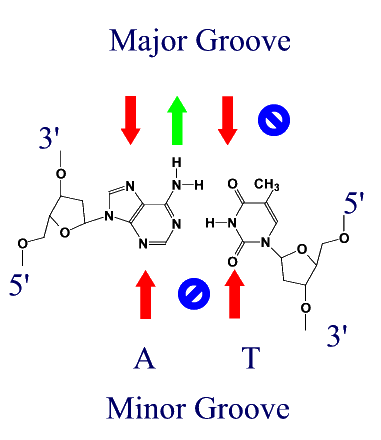
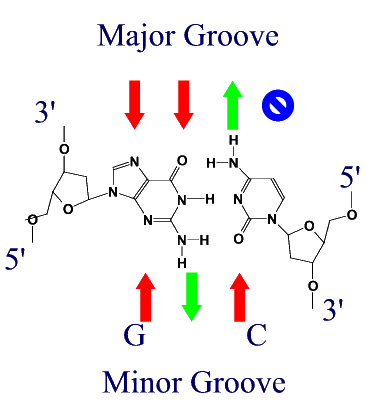
Atomic Interactions between Proteins and DNAProteins bind to DNA through the same forces that hold strands of DNA together, namely H-bonds and ionic interactions. Different amino acids contain H-bond donors and acceptors, and charged residues on their side chains. DNA binding proteins tend to bind in the major groove of DNA for two reasons.
(Red arrows indicate H-bond acceptors and Green arrows H-bond donors) DNA binding motifsDNA binding proteins adopt a set of common three-dimensional structures called motifs. Within these three dimensional motifs different proteins will have different specific amino acids that will recognize specific H-bond donor and acceptor sites on DNA. In this way different DNA binding proteins can recognize very specific DNA sequences. Examples of the most common motifs are shown below.
|
|||