|
Current Level |
||||||||||||||||||
|
|
||||||||||||||||||
|
Previous Level |
||||||||||||||||||
|
|
||||||||||||||||||
|
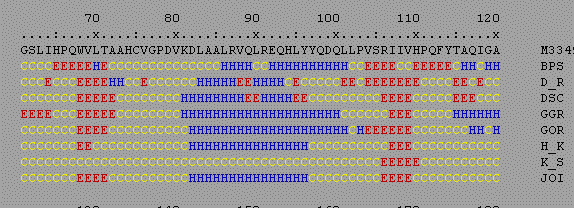
Several programs are available to predict protein secondary structures (alpha helicies and beta sheets). These programs use different algorithms to make these predictions, but often arrive at similar conclusions. Such information can be useful in predicting the approximate structure of a protein or a particular region of interest in a protein. The program PELE on BIOLOGY WORKBENCH is very useful in that it predicts secondary structure using eight different programs and aligns their results. This allows you to rapidly compare the results from several models. Select the amino acid sequence you wish to analyze in the Protein Tools section of Biology Workbench. Then select PELE. You will get results similar to those below.
H = alpha helix, E = beta sheet, T = beta turn, C = random coil As you can see in this example, most of the models predict an alpha helix from residues 83-96 and beta sheets from 68-72 and 106-109. The region from 115-120 gives different results with the different models, and apprears to be more difficult to predict. By comparing the results from several programs you can judge how confident you are in the predicted structure.
|
|
|
|