|
Current Level |
||||||||||||||||||
|
|
||||||||||||||||||
|
Previous Level |
||||||||||||||||||
|
|
||||||||||||||||||
|
What is a primer? A primer is a short synthetic oligonucleotide which is used in many molecular techniques from PCR to DNA sequencing. These primers are designed to have a sequence which is the reverse complement of a region of template or target DNA to which we wish the primer to anneal.
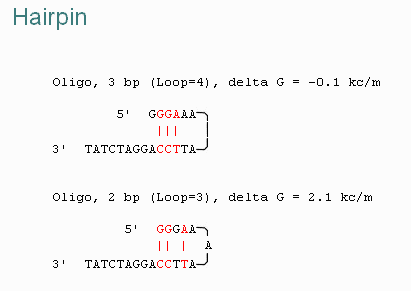
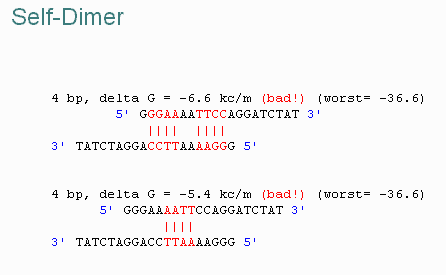
The programs will calculate both the Tm of the primers, as well as any undesireable pairings of primers. When primers form hairpin loops or dimers less primer is available for the desired reaction. For example...
Some thoughts on designing primers. 1. primers should be 17-28 bases in length; Also keep in mind that most oligonucleotide synthesis reactions are only 98% efficient. This means that each time a base is added, only 98% of the oligos will receive the base. This is not often critical with shorter oligos, but as length increases, so does the probability that a primer will be missing a base. This is very important in mutagenesis or cloning reactions. Purification by HPLC or PAGE is recommended in some cases.
Designing Degenerate Oligonucleotides. A group of degenerate oligonucleotides contain related sequences with differences at specific locations. These are used simultaneously in the hope that one of the sequences of the oligonucleotides will be perfectly complementary to a target DNA sequence. One common use of degenerate oligonucleotides is when the amino acid sequence of a protein is known. One can reverse translate this sequence to determine all of the possible nucleotide sequences that could encode that amino acid sequence. A set of degenerate oligonucleotides would then be produced matching those DNA sequences. The following link will take you to a program that will perform a reverse translation. http://arbl.cvmbs.colostate.edu/molkit/rtranslate/ For example, the amino acid sequence shown in purple below could be encoded by the following codons.
One could then select the 14 base sequence (in blue) to generate a smaller set of degenerate oligonucleotides. Each oligonucleotide in the set would have one base changed at a time (shown in purple below). A total of 32 unique oligonucleotides would be generated.
When ordering degenerate oligonucleotides, you just let the company know that you want a mixture of nucleotides added at a specific position using the code below. By adding the mixture, oligos will incorporate one of the bases, leading to a mixture of oligonucleotides.
|
|||||||||||||||||||||||||||||||||||||||
|
|
|
|
|