|
Current Level |
||||||||||||||||||
|
|
||||||||||||||||||
|
Previous Level |
||||||||||||||||||
|
|
||||||||||||||||||
|
Translation and Open Reading Frame Search Regions of DNA that encode proteins are first transcribed into messenger RNA and then translated into protein. By examining the DNA sequence alone we can determine the sequence of amino acids that will appear in the final protein. In translation codons of three nucleotides determine which amino acid will be added next in the growing protein chain. It is important then to decide which nucleotide to start translation, and when to stop, this is called an open reading frame. Once a gene has been sequenced it is important to determine the correct open reading frame (ORF). Every region of DNA has six possible reading frames, three in each direction. The reading frame that is used determines which amino acids will be encoded by a gene. Typically only one reading frame is used in translating a gene (in eukaryotes), and this is often the longest open reading frame. Once the open reading frame is known the DNA sequence can be translated into its corresponding amino acid sequence. An open reading frame starts with an atg (Met) in most species and ends with a stop codon (taa, tag or tga). For example, the following sequence of DNA can be read in six reading frames. Three in the forward and three in the reverse direction. The three reading frames in the forward direction are shown with the translated amino acids below each DNA seqeunce. Frame 1 starts with the "a", Frame 2 with the "t" and Frame 3 with the "g". Stop codons are indicated by an "*" in the protein sequence. The longest ORF is in Frame 1. Practice Problems and a Translation Lecture Tutorial are located in the Theory Section.
DNA Translation. To translate a DNA sequence, we use the program called SIXFRAME on the Biology Workbench. Or you can visit the site directly http://searchlauncher.bcm.tmc.edu/seq-util/Options/sixframe.html
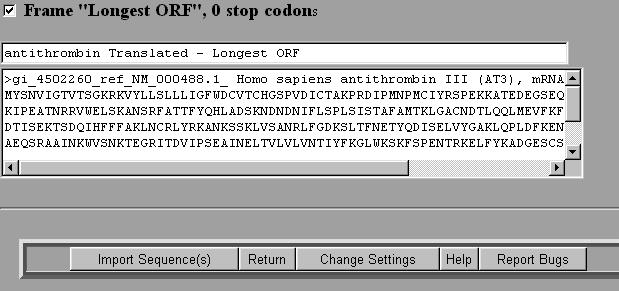
2. Select Nucleic Tools and then Select the sequence that you wish to translate by checking the box next to the file. 3. Select SIXFRAME and then select Run. 4. You will be given a box asking you to select various parameters. The default is to translate all six reading frames of the entire DNA sequence. If you select ``Show longest open reading frame" the program will automatically select the longest reading frame starting with a start codon (ATG) and ending with a stop codon (TAA, TAG or TGA). This is very handy. 5. Once the parameters are chosen select Submit.
Interpretation of Results
|
|
|
|